Research Summary
1. About “Molecular Movies”
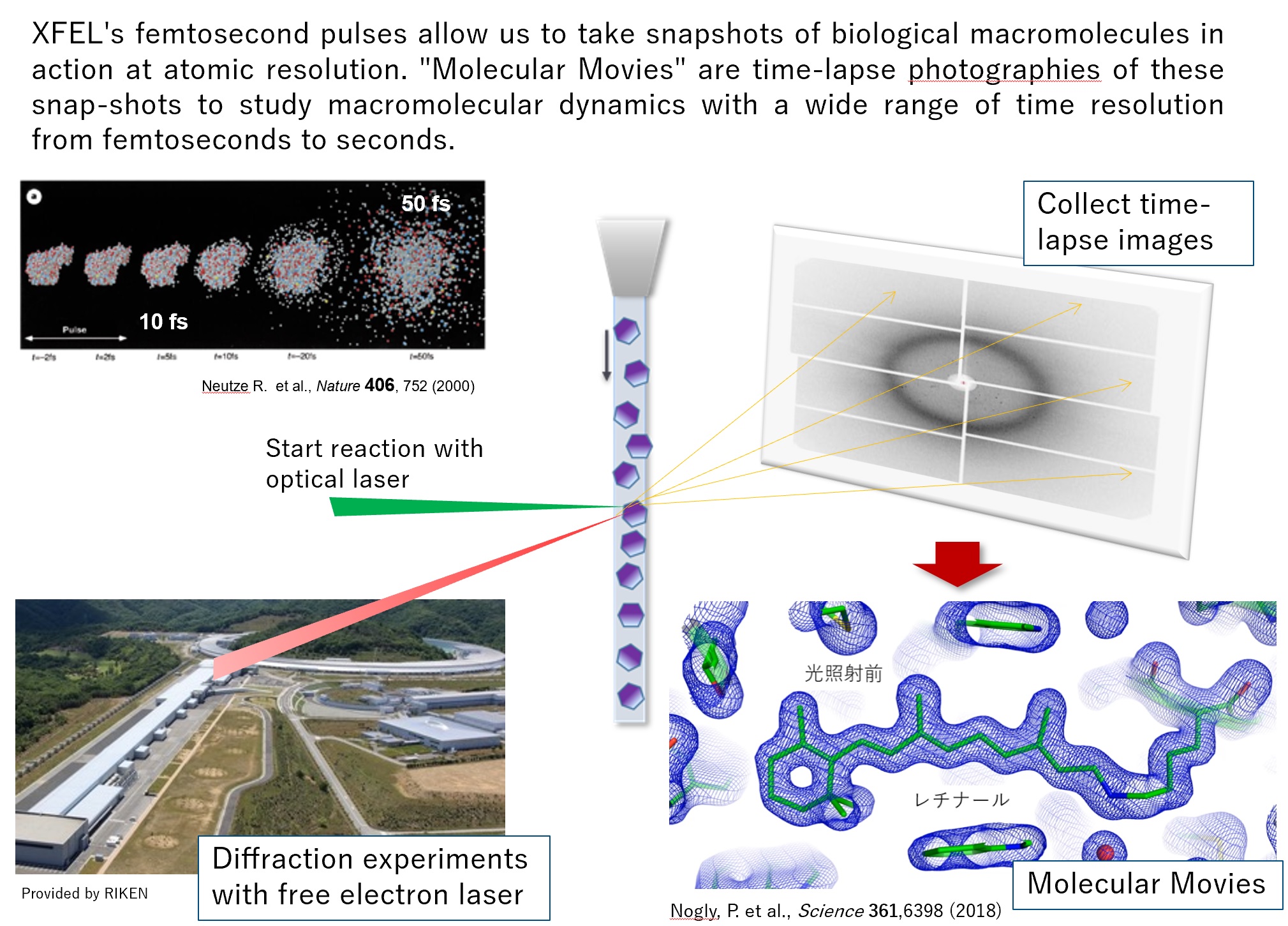
In Molecular Movies, we can take a snapshot of the structure after a certain time (with a wide range of time resolution from femtoseconds to seconds) from the start of the reaction. By combining a series of time-lapse images with different delay times, we have been able to capture the movement of molecules as a movie.
2. Aim of the the project
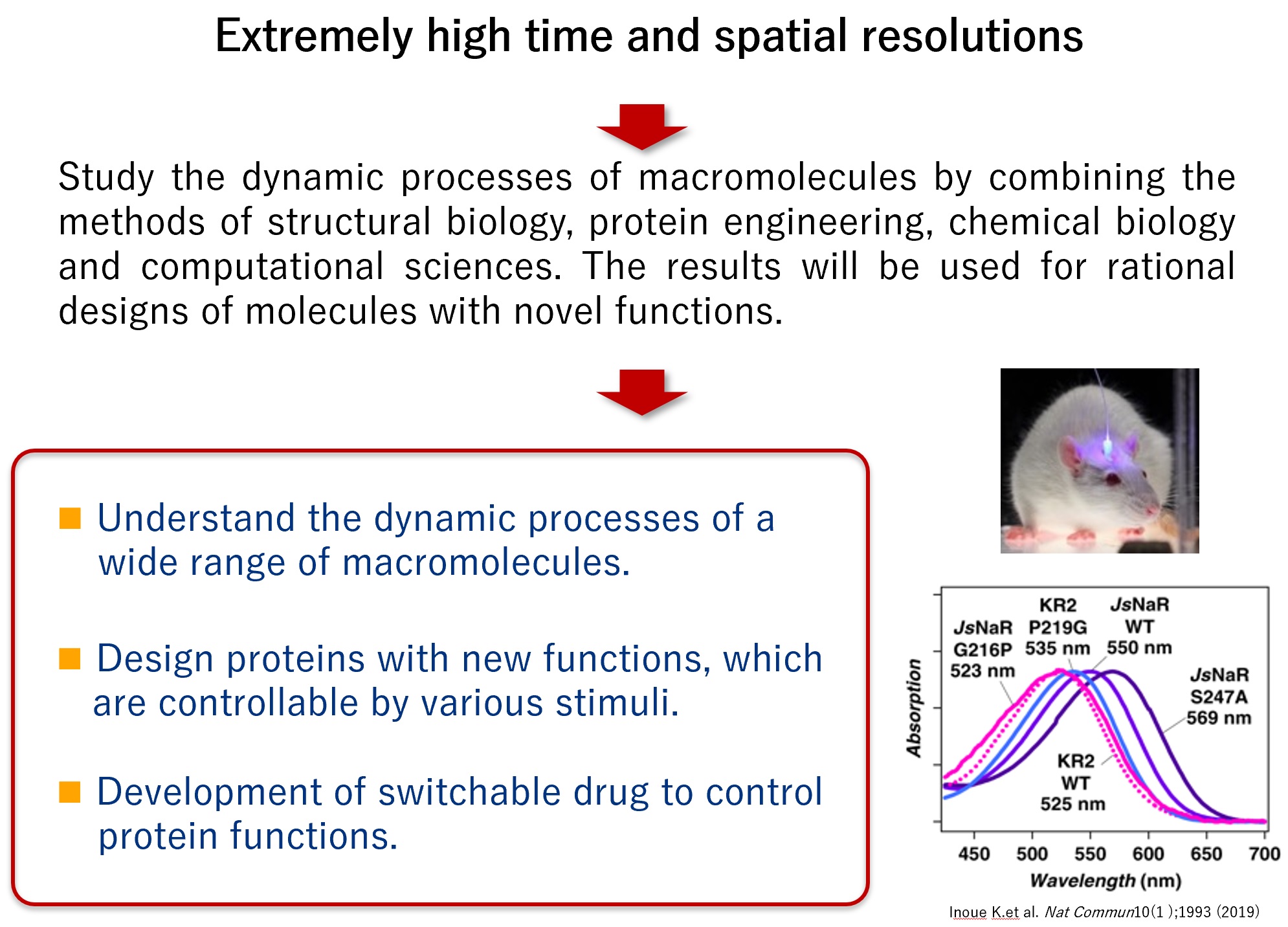
In our study, we will elucidate the dynamic mechanism of biomolecules by utilizing Molecular Movies method. Using the results, we aim to develop proteins that can be switched by various stimuli and substances that can control proteins. To this end, we make full use of computational science, protein engineering, and chemical biology as well as structural biology.
3. Early results
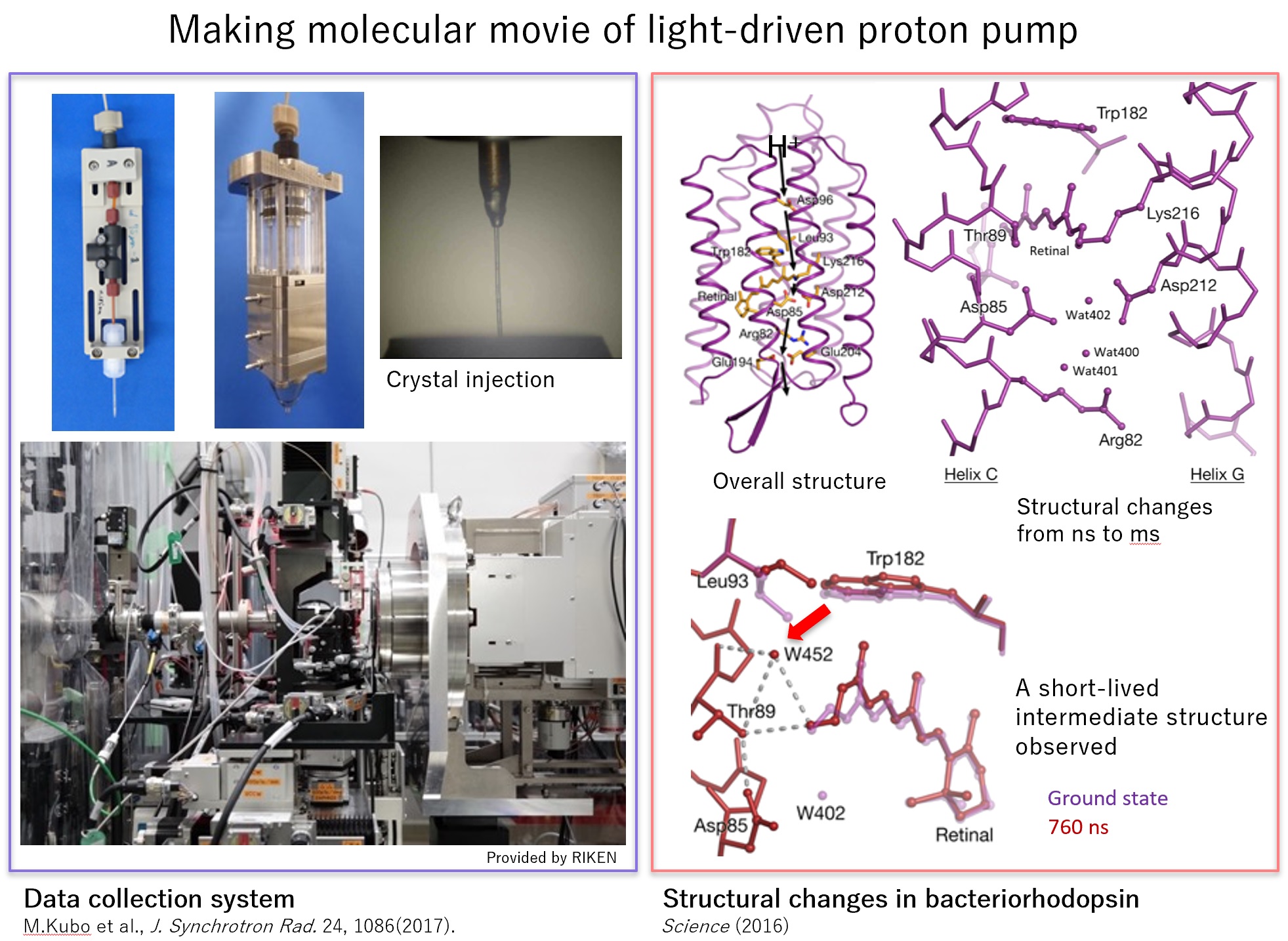
We have developed the experimental system based on Serial Femtosecond Crystallography (SFX) at SACLA. First, as a demonstration experiment, we succeeded in capturing structural changes from 20ns to 2ms after exciting bacteriorhodopsin with light.
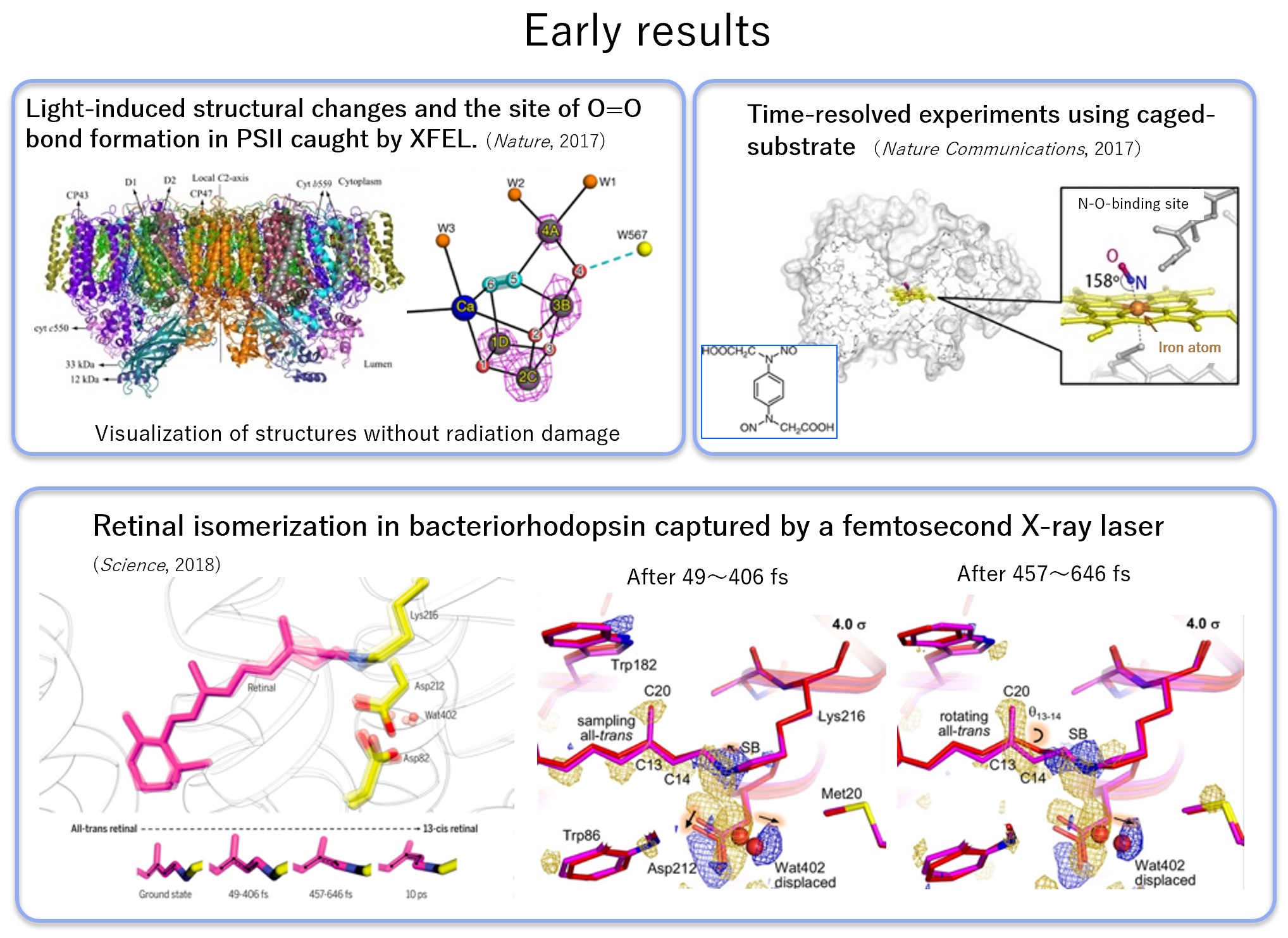
Next, we adapted the developed system to Photosystem II. And we succeeded in capturing an important intermediate for the enzyme to generate oxygen from water molecules using the energy of light. We also succeeded in capturing complexes with nitric oxide reductase substrates using caged compounds, aiming to expand the application to proteins that are not sensitive to light. We analyzed bacteriorhodopsin with faster temporal resolution, observed the rotation of retinal binding from fs to ps, and elucidated the movement of nearby water molecules and protein side chains. Reference
- A three-dimensional movie of structural changes in bacteriorhodopsin.
Nango E. et al. Science, 354(6319):1552-1557(2016)
DOI : 10.1126/science.aah3497 - Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Suga M. et al. Nature, 543:131–135(2017)
DOI : http://doi.org/10.1038/nature21400 - Capturing an Initial Intermediate during the P450nor Enzymatic Reaction using Time-Resolved XFEL Crystallography and Caged-Substrate
Tosha T. et al. Nature Communications, 8:1585(2017)
DOI :10.1038/s41467-017-01702-1 - Retinal isomerization in bacteriorhodopsin captured by a femtosecond X-ray laser
Nogly P. et al. Science, 361(6398):eaat0094(2018)
DOI: 10.1126/science.aat0094
We have posted on equipment development on the SACLA-SFX website.
4. Objectives
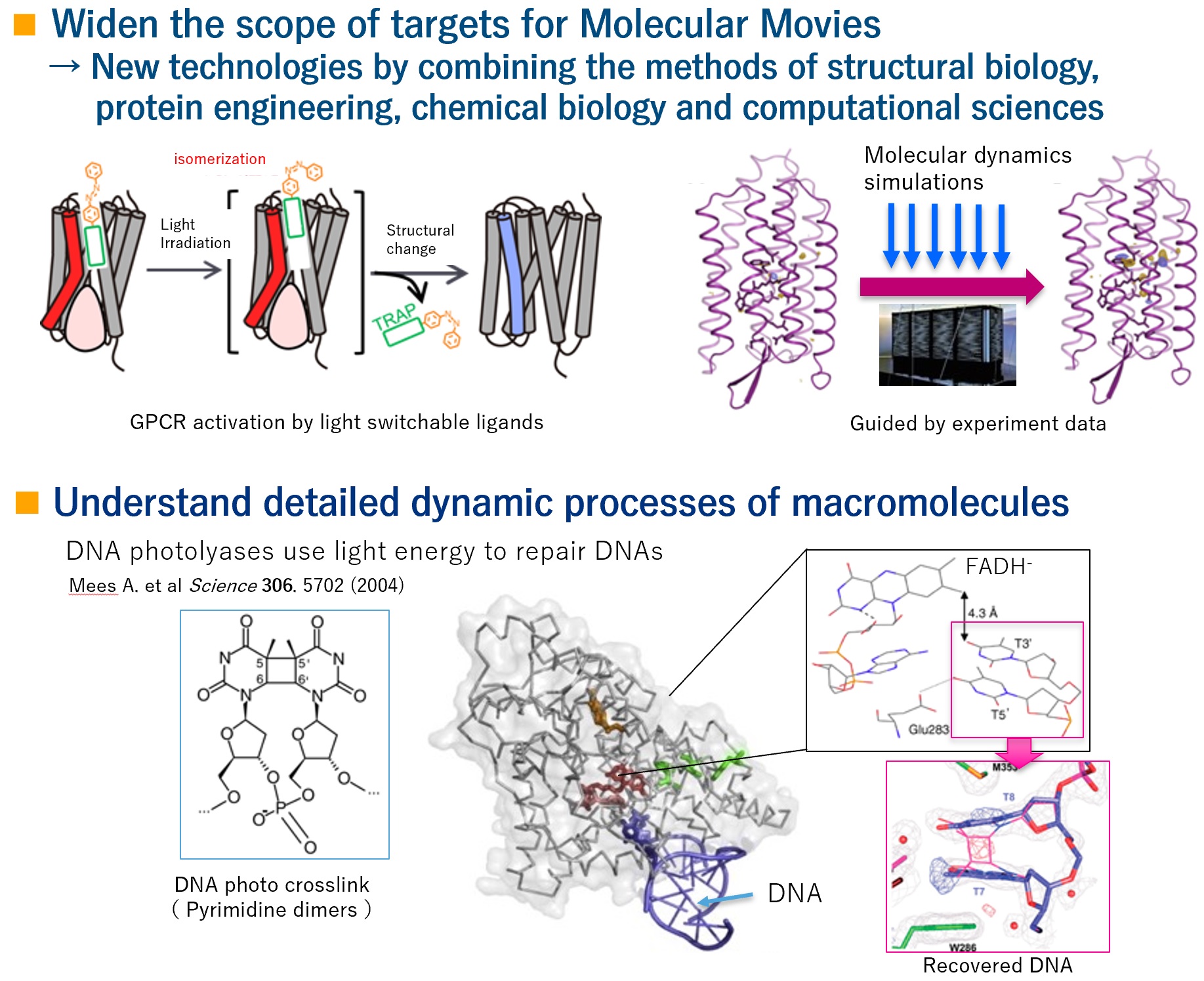
Our primary goal is to broaden the base of Molecular Movies. So far, Molecular Movies has been mainly applied to light-sensitive proteins. To broaden this application to more biologically interesting targets, we will build a mechanism to switch molecules with light using protein engineering and chemical biology. We will also pursue the synchronization methods other than light by developing measuring equipment. We interpret the obtained results more quantitatively and theoretically using computational science. Based on that, we will deepen our understanding of the dynamic mechanism of various biomolecules.
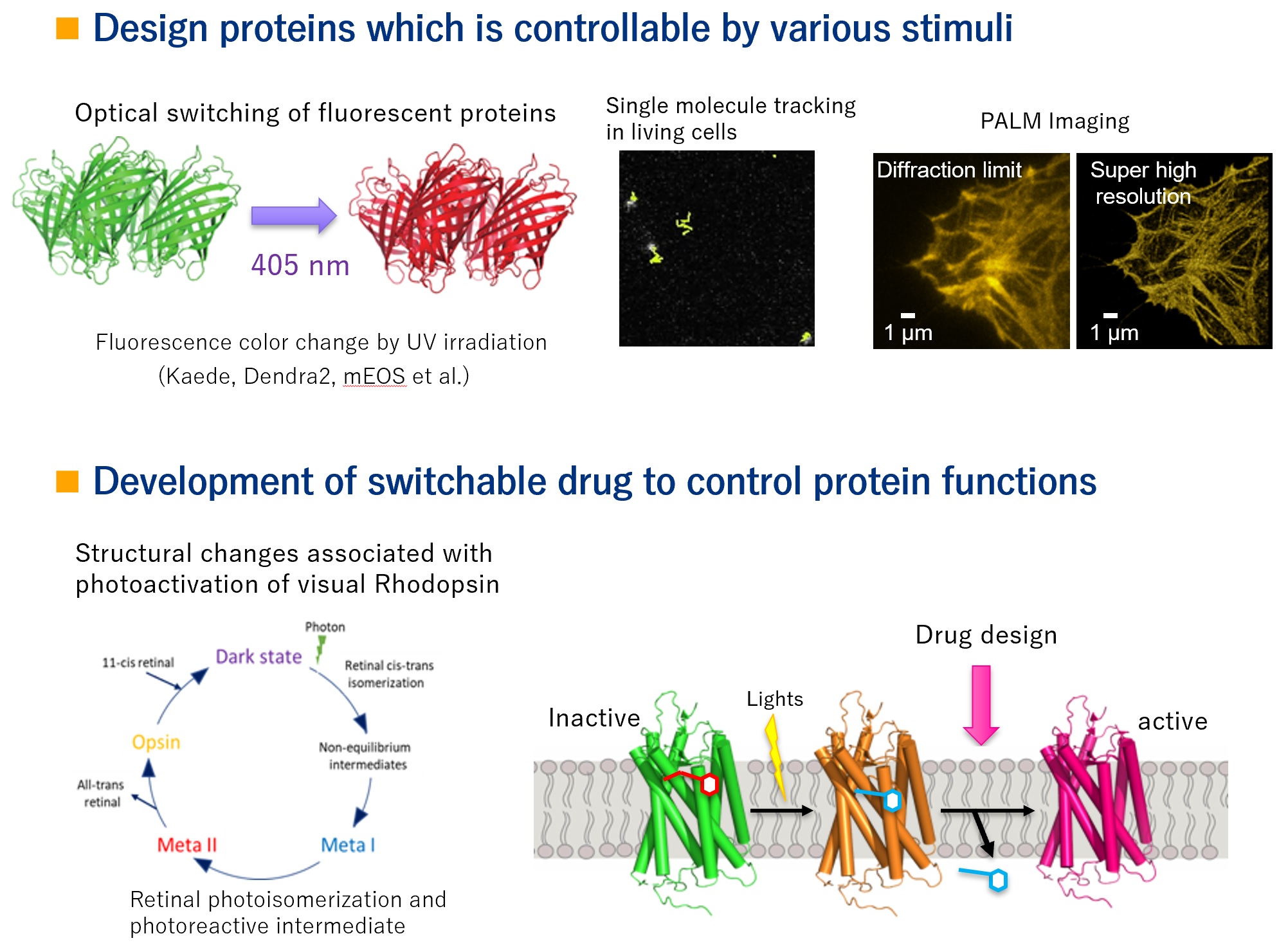
We are not only observing the dynamic structure of biomolecules, but we want to link it to the development of new functional molecules. For example, we would like to develop new fluorescent and luminescent proteins used for imaging and optogenetics, and functional drugs that can be used for imaging and photopharmacology.
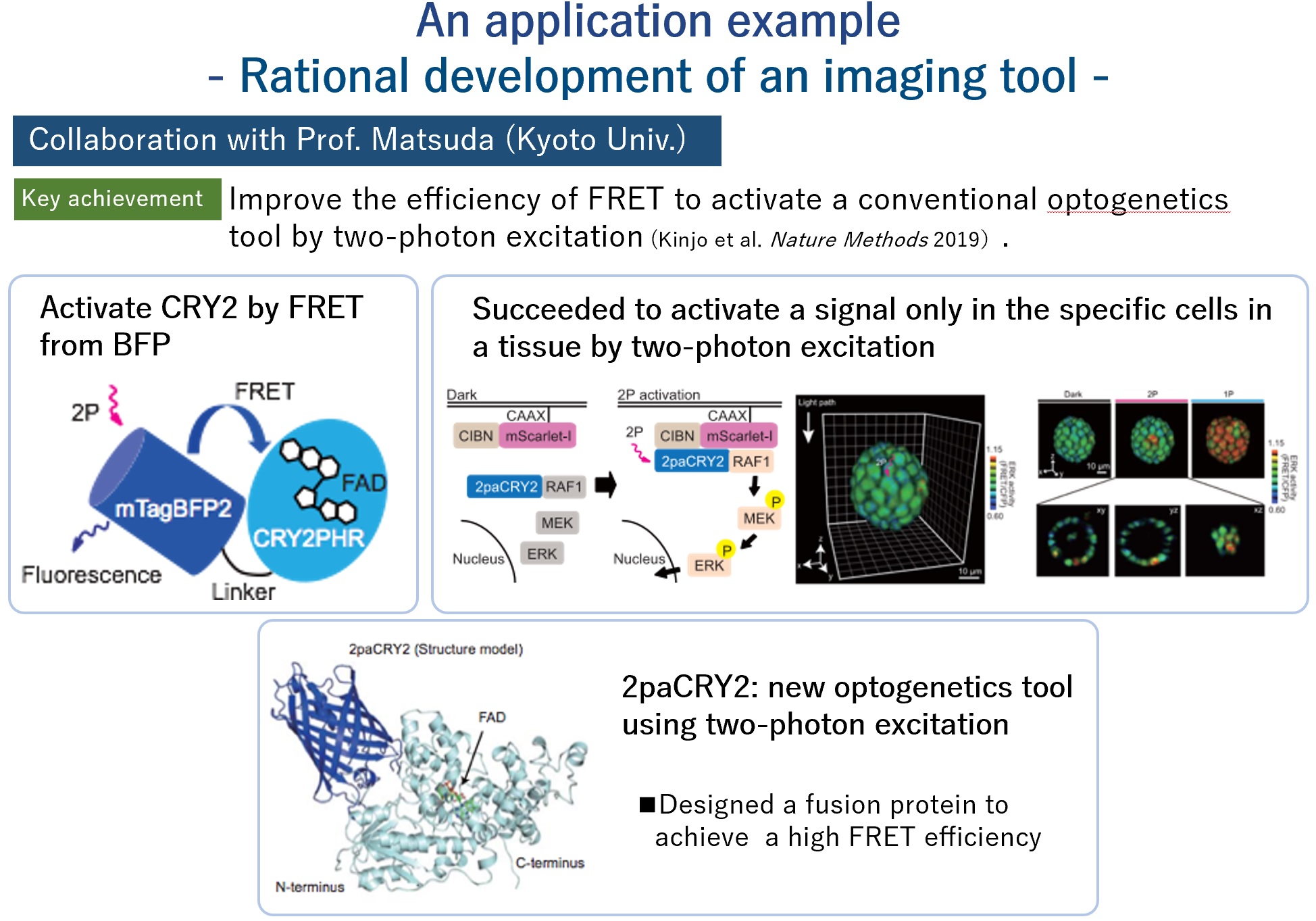
We have already started and succeeded in achieving high efficiency of conventional optogenetics tools from protein structure information. (Joint research with Prof. M. Matsuda (Kyoto Univ.), Kinjo et al. Nature Methods 2019) In this way, we would like to utilize the dynamic information obtained from the Molecular Movies method to develop new tools.

